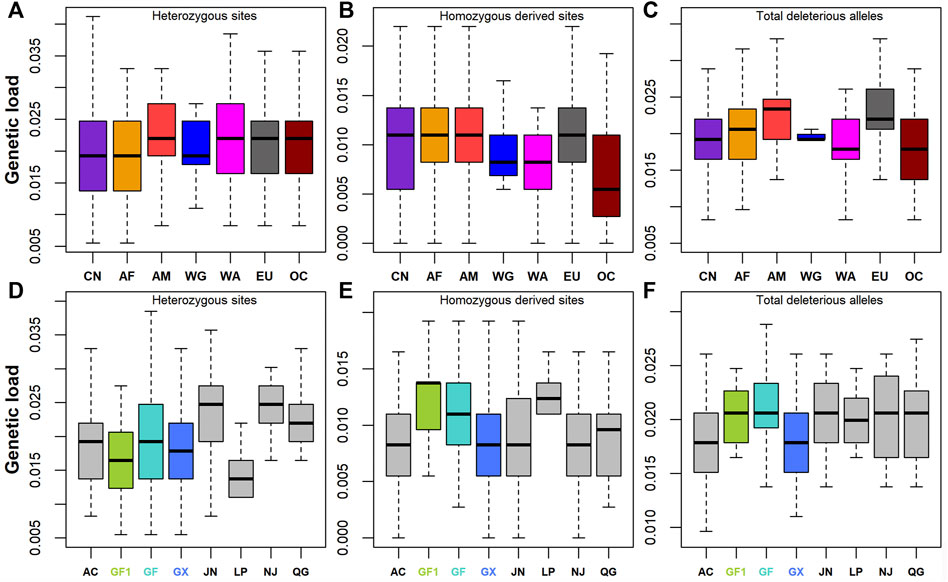
Frontiers | Genome-Wide Estimates of Runs of Homozygosity, Heterozygosity, and Genetic Load in Two Chinese Indigenous Goat Breeds
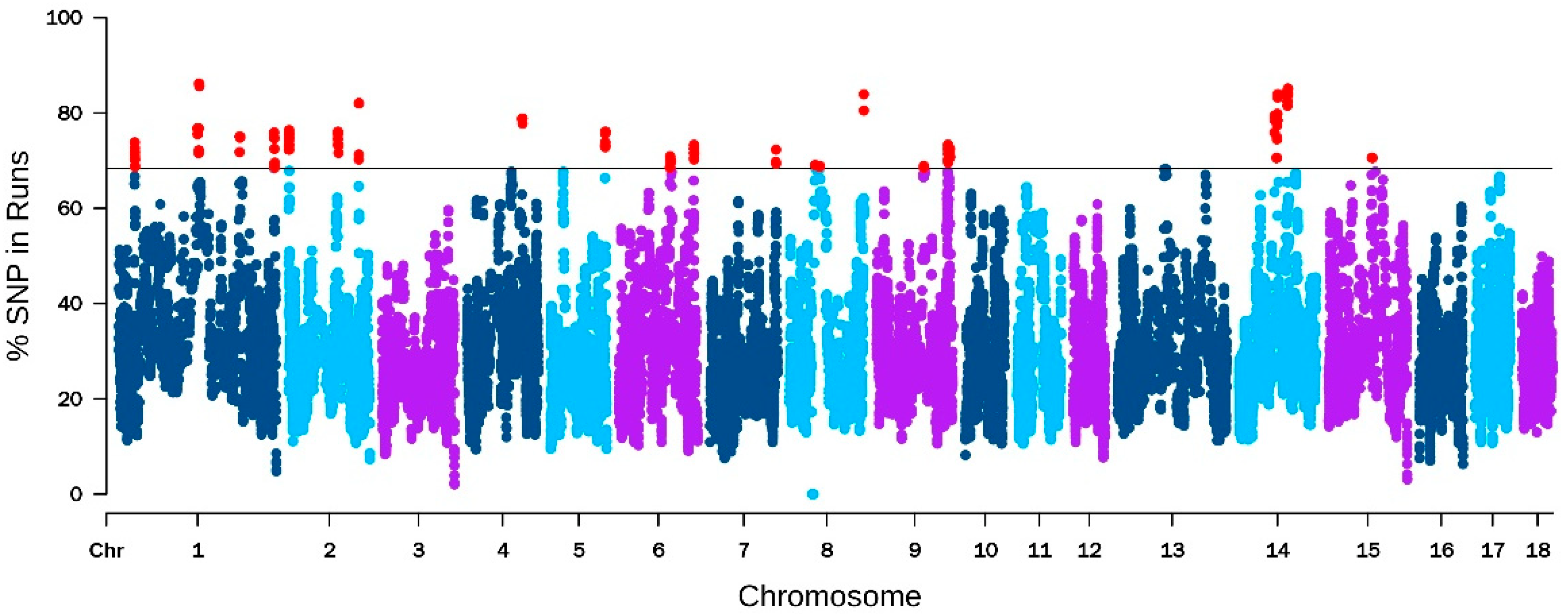
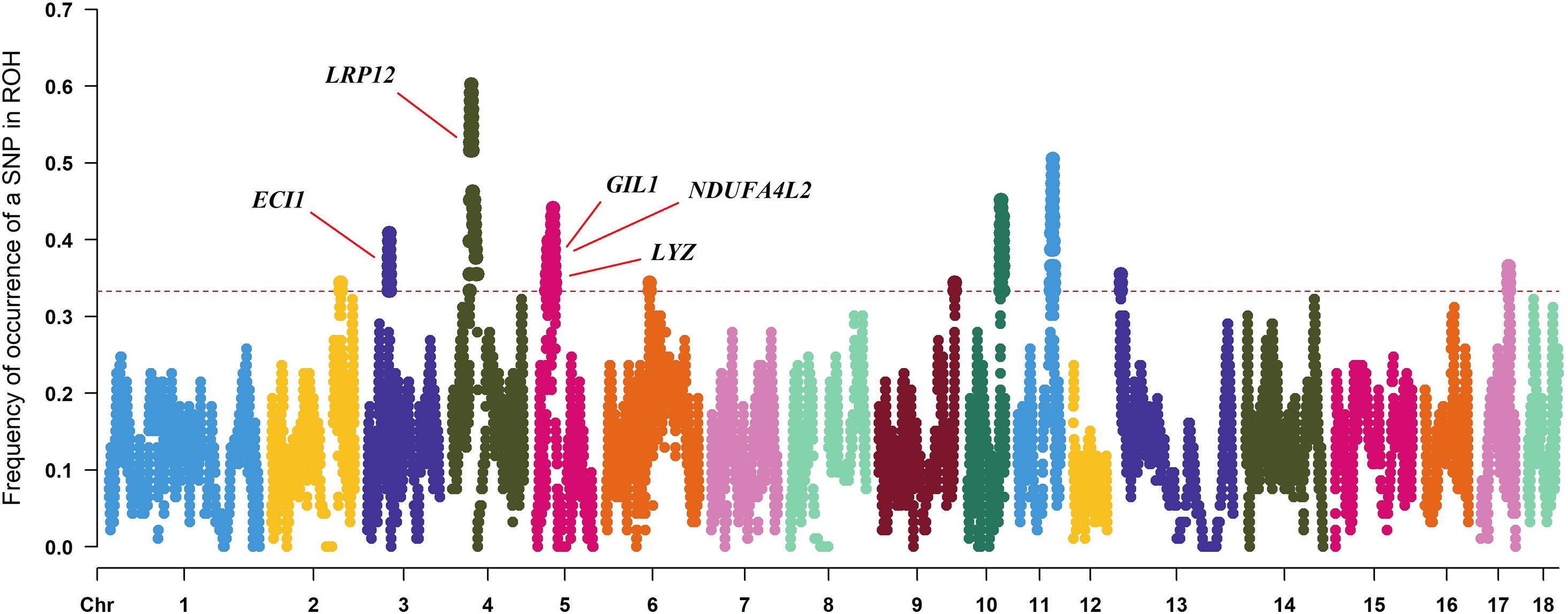
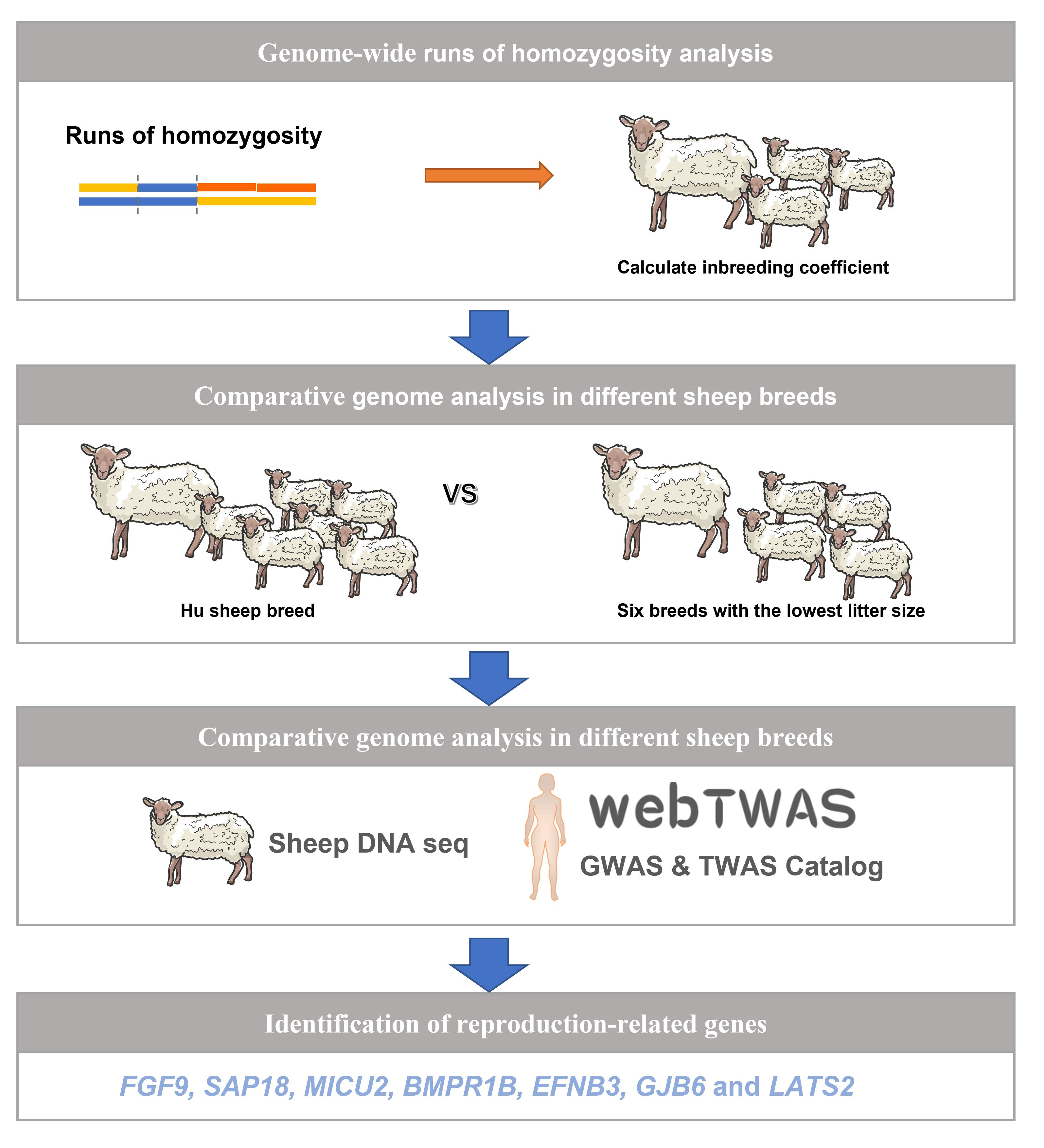
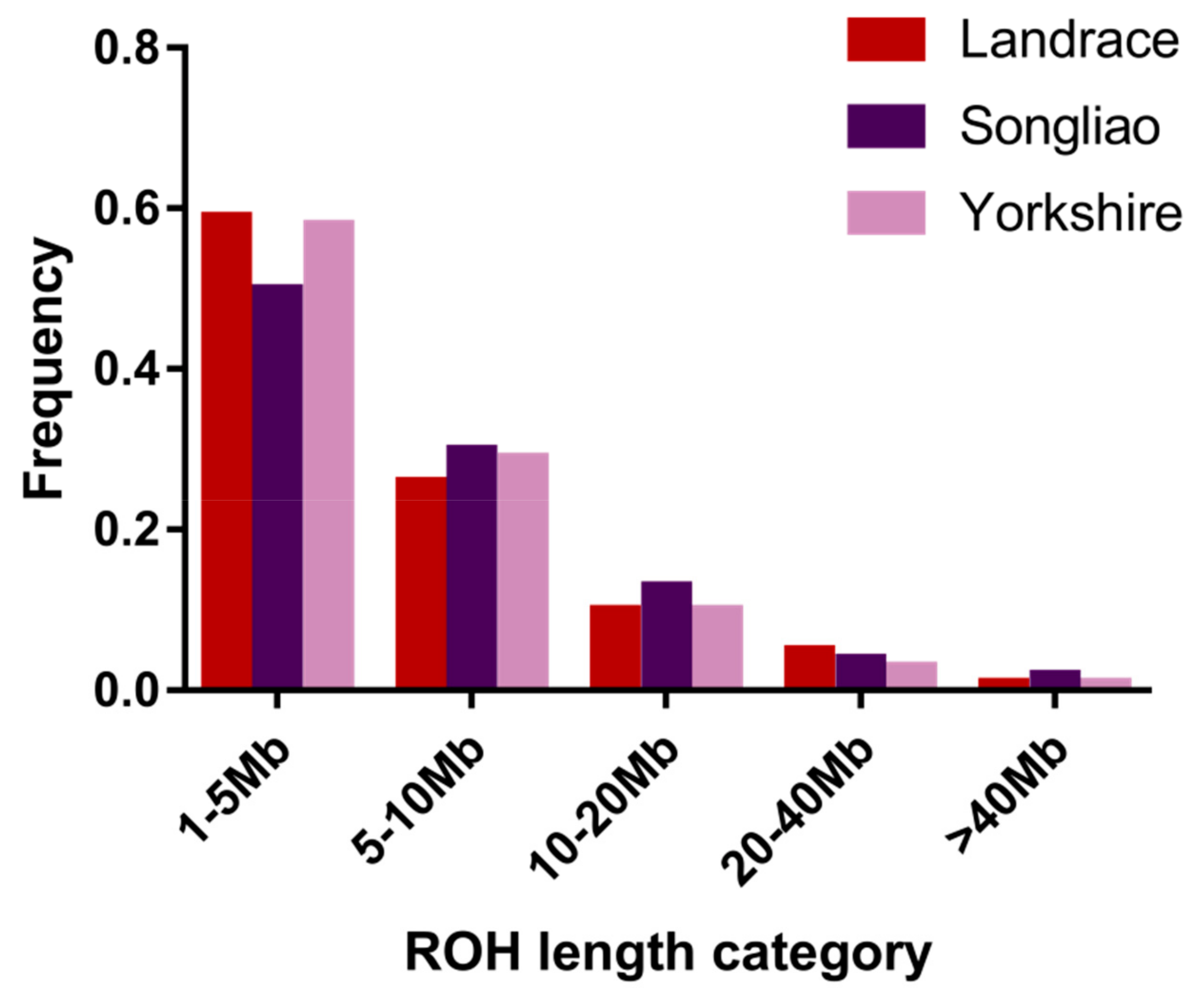
Genes | Free Full-Text | Genome-Wide Association Studies and Runs of Homozygosity to Identify Reproduction-Related Genes in Yorkshire Pig Population
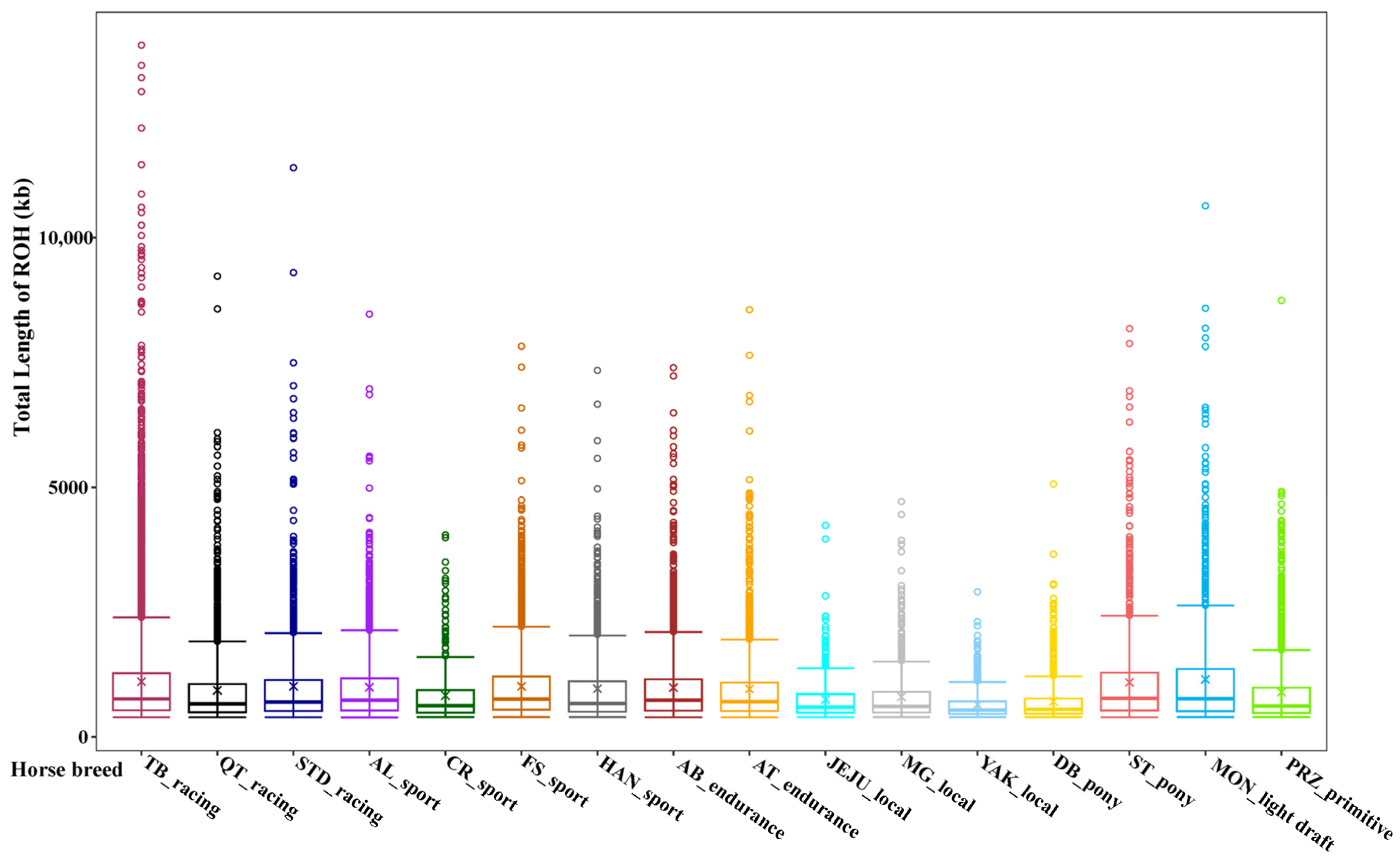
Genes | Free Full-Text | Genome-Wide Assessment of Runs of Homozygosity by Whole-Genome Sequencing in Diverse Horse Breeds Worldwide
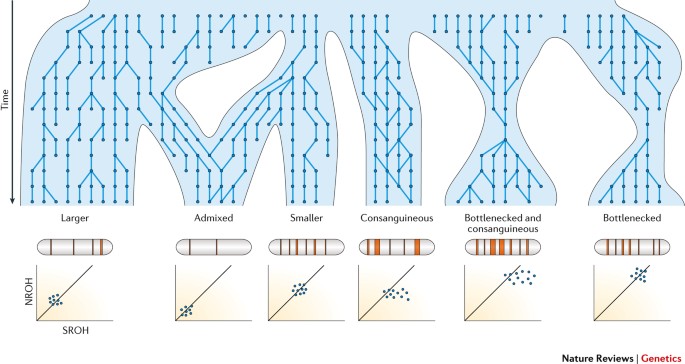
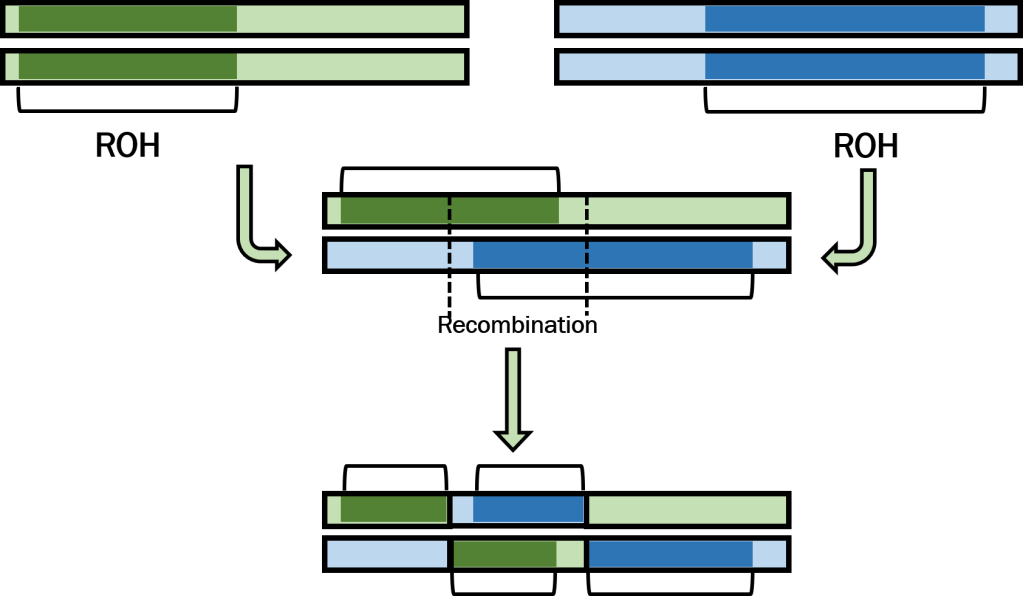
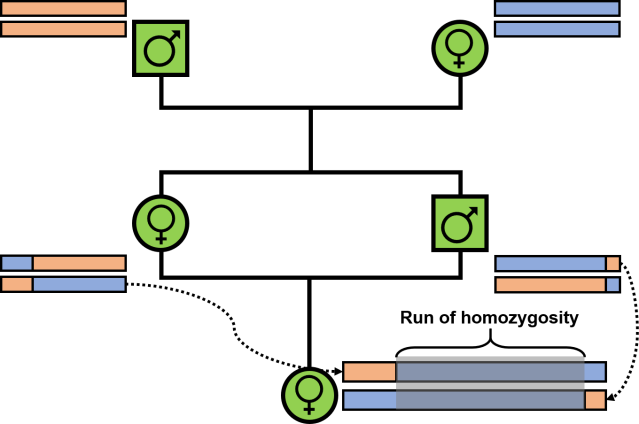
Runs of homozygosity: windows into population history and trait architecture | Nature Reviews Genetics

John Novembre on X: "You'd think calling runs of homozygosity from pseudohaploid data would be impossible. It's not. The wonders and power of LD can make it go. Really happy to see

The lengths and number of runs of homozygosity (ROH) segments across... | Download Scientific Diagram

Discovery of runs-of-homozygosity diplotype clusters and their associations with diseases in UK Biobank | medRxiv
![PDF] Runs of homozygosity: windows into population history and trait architecture by Francisco C. Ceballos, Peter K. Joshi, David W. Clark, Michèle Ramsay, James F. Wilson · 10.1038/nrg.2017.109 · OA.mg PDF] Runs of homozygosity: windows into population history and trait architecture by Francisco C. Ceballos, Peter K. Joshi, David W. Clark, Michèle Ramsay, James F. Wilson · 10.1038/nrg.2017.109 · OA.mg](https://og.oa.mg/Runs%20of%20homozygosity%3A%20windows%20into%20population%20history%20and%20trait%20architecture.png?author=%20Francisco%20C.%20Ceballos,%20Peter%20K.%20Joshi,%20David%20W.%20Clark,%20Mich%C3%A8le%20Ramsay,%20James%20F.%20Wilson)
PDF] Runs of homozygosity: windows into population history and trait architecture by Francisco C. Ceballos, Peter K. Joshi, David W. Clark, Michèle Ramsay, James F. Wilson · 10.1038/nrg.2017.109 · OA.mg

Box plot of the distribution of runs of homozygosity for each breed... | Download Scientific Diagram
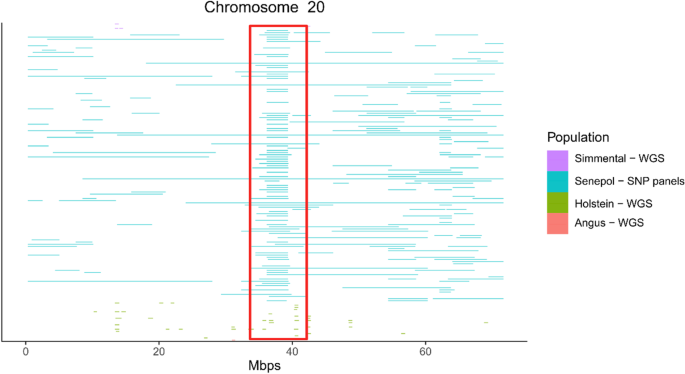
Characterization of runs of homozygosity, heterozygosity-enriched regions, and population structure in cattle populations selected for different breeding goals | BMC Genomics | Full Text
Animals | Free Full-Text | Genome-Wide Scan for Runs of Homozygosity Identifies Candidate Genes in Three Pig Breeds

How to study runs of homozygosity using PLINK? A guide for analyzing medium density SNP data in livestock and pet species | BMC Genomics | Full Text
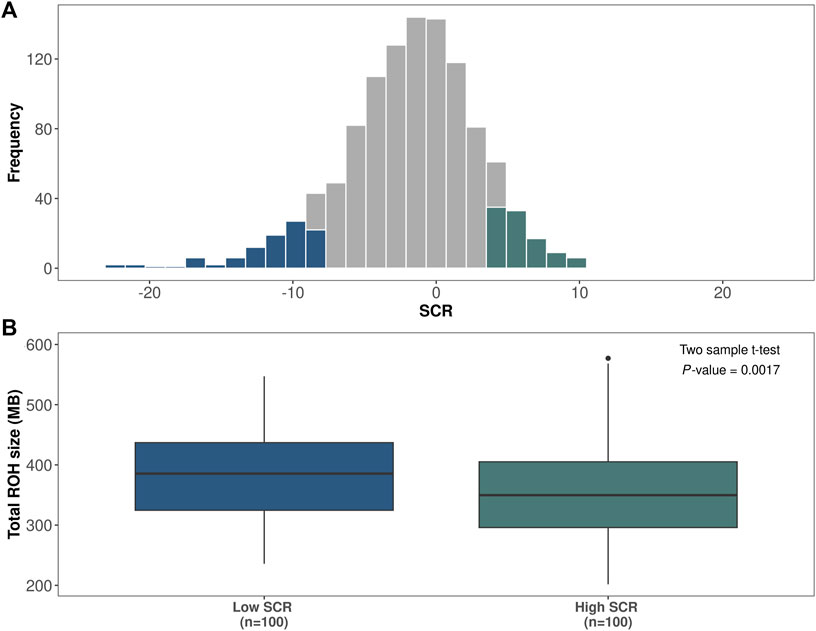
Frontiers | Identification of runs of homozygosity associated with male fertility in Italian Brown Swiss cattle

The pattern of runs of homozygosity and genomic inbreeding in world-wide sheep populations - ScienceDirect
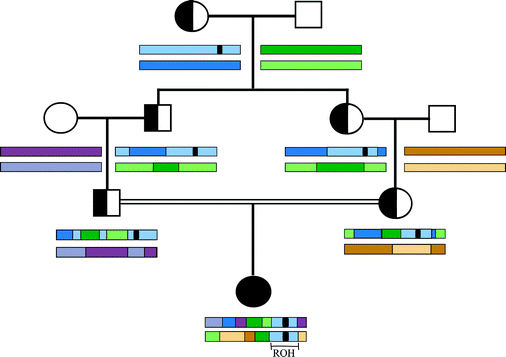
Evaluating Runs of Homozygosity in Exome Sequencing Data - Utility in Disease Inheritance Model Selection and Variant Filtering | SpringerLink

An example of a run of homozygosity of 10 SNP (within dotted square)... | Download Scientific Diagram

Runs of homozygosity (ROH) variation among individuals and across the... | Download Scientific Diagram

Global distribution of long runs of homozygosity. The average total... | Download Scientific Diagram
![PDF] Runs of homozygosity for autozygosity estimation and genomic analysis in production animals | Semantic Scholar PDF] Runs of homozygosity for autozygosity estimation and genomic analysis in production animals | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/66d2391f269bc27494df33b7335ec6fcb44904cb/4-Figure1-1.png)